mutation from threonine to what amino acid make phosphorylation
Abstract
Serine is the simply amino acrid that is encoded by two disjoint codon sets (TCN & AGY) so that a tandem substitution of ii nucleotides is required to switch betwixt the two sets. We show that these codon sets underlie distinct substitution patterns at positions subject to purifying and diversifying selections. We found that in humans, positions that are conserved among ~100 vertebrates, and thus subjected to purifying selection, are enriched for substitutions involving serine (TCN, denoted South′), proline, and alanine, (S′PA). In contrast, the less conserved positions are enriched for serine encoded with AGY codons (denoted Southward″), glycine and asparagine, (GS″Due north). We tested this phenomenon in the HIV envelope glycoprotein (gp120), and the V-cistron that encodes B-prison cell receptors/antibodies. These fast evolving proteins both have hypervariable positions, which are under diversifying selection, closely next to highly conserved structural regions. In both instances, nosotros identified an reverse abundance of 2 groups of serine substitutions, with enrichment of South′PA in the conserved positions, and GS″N in the hypervariable regions. Finally, nosotros analyzed the substitutions across lx,000 private human exomes to prove that, when serine has a specific functional constraint of phosphorylation capability, Southward′ codons are 32-folds less prone than S″ to substitutions to Threonine or Tyrosine that could potentially retain the phosphorylation site chapters. Combined, our results, that encompass evolutionary signals at different temporal scales, demonstrate that through its encoding by two codon sets, serine allows for the beingness of alternating commutation patterns inside positions of functional maintenance versus sites of rapid diversification.
Introduction
Due to the redundancy of the genetic code almost amino acids are encoded by more one codon. In most cases the codons encoding for a specific amino acid differ simply past a single nucleotide (typically the third one). Nevertheless, serine is unique amid the 20 amino acids in that it is encoded by two disjoint sets of codons that crave a tandem exchange of two nucleotides to switch between the ii sets, one comprised of the codons TCT, TCC, TCA and TCG (TCN, denoted set S′) and the other gear up includes but 2 codons - AGT and AGC (AGY, denoted set S′′)1. The ii sets of codons encoding serine cannot exist connected through a single signal mutation1. The serine'south codon structure allows it to participate in two split up substitution patterns, each of which conserves different amino acid characteristics. The South′′ unmarried mutation transition commutation category includes the amino acids serine, in addition to transition neighbors2,3,four glycine (G), and asparagine (N). In contrast, the S′ unmarried mutation transition commutation category includes serine, proline (P), and alanine (A)3,5,6 (Fig. 1). The substitution of S′ to amino acids P and A occur past a unmarried mutation at the outset codon position. The commutation from serine to threonine (T) is non included either of the exchange sets, every bit it tin can be a event of a unmarried nucleotide modify from either S′ of S″. While the amino acid substitution derived from Due south′ are enriched in conserved β-turnsthree,vi,7, those derived from S″ are generally of a neutral hydrophobicity (i.e., amino acids that are neither strongly hydrophobic or hydrophilic)iii,6.
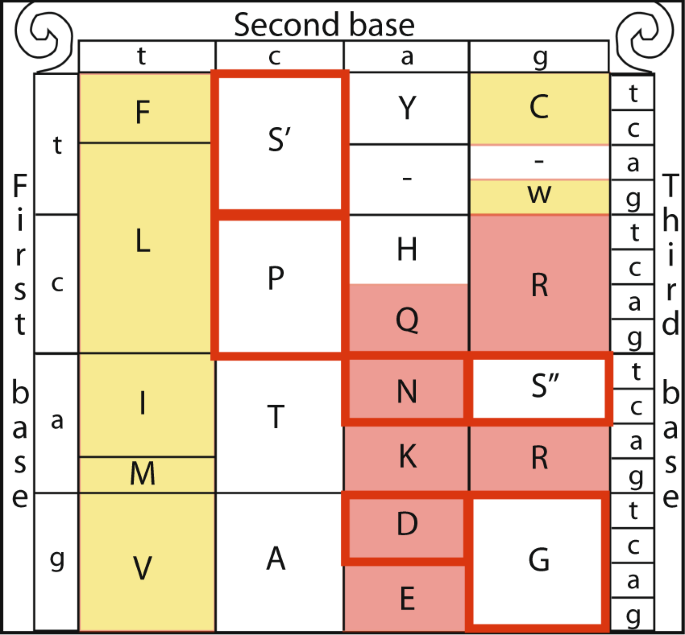
The genetic code annotated for four types of amino acid characteristics. Hydrophobic (yellow fill up), Neutral hydrophobicity (white), Hydrophilic (red) and β−Turn (red frame)iii,6,7.
We tested the usage of the two patterns of serine substitutions in different evolutionary contexts. Substitution patterns were studied across 99 vertebrates in HIV envelope glycoprotein, and in humans for the germline and somatic diversification of the allowed B jail cell receptor repertoire. Nosotros demonstrated that the special form of the codon back-up encoding serine underlies serine's participation in two distinct exchange patterns. One of which (amid the amino acids – GS″Northward) is more abundant in sequence positions subject to positive, diversifying option, while the other (amongst - Southward′PA) is more than abundant at highly constrained positions, subject to strong purifying pick. Additionally, we detected that substitutions of the S′ set associated with phosphorylation sites that are under a stronger constraint relative to that of the S″ gear up. It therefore appears that the genetic code has evolved to specifically allow serine to participate in two types of selective regimes, ane that complies with potent constraint and another separated regime for diversification.
Results
Serine displays a natural segmentation to 2 codon sets with singled-out exchange patterns
We hypothesized that each of the individual serine codon sets had its own preferred substitutions, and that these two groups of serine substitutions would inhabit dissimilar position and backdrop in protein sequence. To characterize the impact of the differential distribution of serine on its genome wide substitution patterns, we reconstructed a new version of the well characterized BLOcks SUbstitution Matrix (BLOSUM)viii (see Methods) that split up the substitution matrix for serine into S′ and South′′. We found that the 2 codon-based serine substitution sets differ in the direction of their substitution scores. The substitution values from Due south′′ to G and Due north are both positive, indicating they occur more ofttimes than expected by adventure, while exchange values from South′ to Grand and N are negative. At the same time, commutation values from S′ to P and A are positive while those from South′′ are not (Fig. 2a).
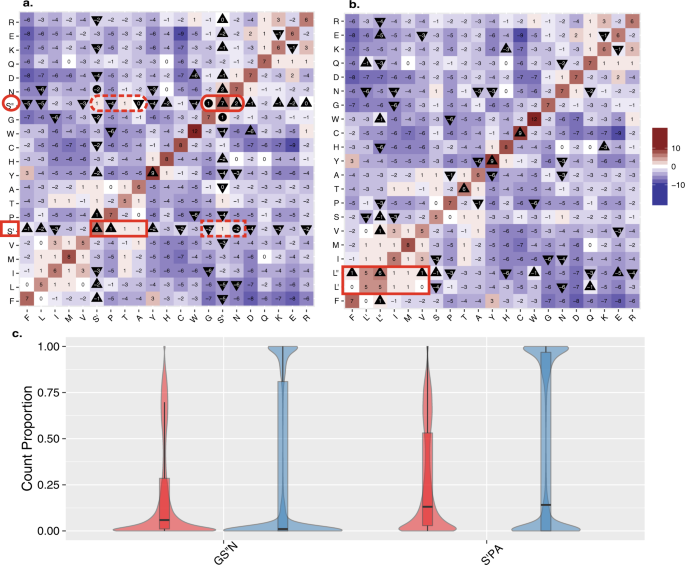
Comparing of codon and amino acid substitutions in the alignment database of 99 vertebrates species. (a) BLOSUM heat map from the UCSC alignments generated while considering substitutions to/from Due south′ and Southward′′ separately. Heat map colored from blue (<−ten) to cherry (>10) with a midpoint slope of white (0). A triangle pointing up designates an increment in value compared to the unmanipulated UCSC based BLOSUM (Sup. Fig. 1), while a triangle pointing down represents a decrease in value. A circle signifies a sign flip (+to - or vice versa) compared to the unmanipulated BLOSUM (Sup. Fig. ane). Annotation that the viable/positive substitutions from S′′ (red oval) are all negative from South′ (cherry-red rectangle broken line), similarly viable/positive substitutions from South′ (ruddy rectangle) are not from S′′(red oval broken line ). T, which is equally reachable from both Due south′ and South″ remains unchanged. (b) The BLOSUM generated by considering substitutions to/from L′ and 50′′ separately (details equally in a). Annotation how no positive substitutions from L′ or 50′′ are negative from the other and all negative substitutions are shared (positive marked in red rectangle). (c) Considering just those positions where serine is meaningfully present (see Methods), we show the fraction of species sequence positions that are GS′′N (two plots on the left) or S′PA (2 plots on the right). For each set up of amino acids, the species sequence positions are split by the measure of multifariousness into positions above the upper quartile of diversity (red) and beneath the lower quartile of multifariousness (blue). Each distribution is represented by a box plot overlaying a violin plot. The black line represents the median value of each grouping.
In accordance with the original BLOSUM, in our reconstructed matrix, synonymous mutations are ever associated with positive values. Withal, nosotros observe that synonymous mutations of S′ or South′′ (i.e. the values on the diagonal) all have values approximately six-fold college than the value reported for the synonymous mutation in which S′ is substituted to Due south′′ or vice versa. The disregarded mutation altitude between the synonymous mutations from S′ to Due south′′, is not considered by the classical BLOSUM. Remarkably, in all cases when we consider the differences between S′ and S′′ substitutions with respect to the substitutions of serine in the original, unmodified version of the BLOSUM matrix, we find that they accept also changed appropriately. Moreover, in the case of substitutions from S′′, several types of substitutions flipped the sign of their values from negative to positive compared to the original BLOSUM (Fig. 2a, Sup. Fig. S1a).
In addition to serine, leucine (L) and arginine (R) are also encoded by half dozen codons which are composed of 2 sets. To check whether the differences of our S′/S′′ matrix from the standard BLOSUM could exist merely attributed to a more detailed clarification of codons, we repeated the analysis and tested the natural split of leucine into CTN (CTT, CTC, CTA, CTG, denoted L′ prepare) and TTR (TTA, TTG, denoted L″ set). For L′ and 50″ we plant no difference in the sign of the commutation scores. The leucine substitution scores change in their extent but not in their tendency for beingness substituted (as reflected past the sign of the substitution values). Furthermore, the synonymous changes of L′ to L′, L′′ to L′′, and the cross sets of L′ to L′′ and L′′ to 50′ all receive approximately equal values (Fig. 2b). We observed the same trend when arginine was tested. Splitting arginine into R′ and R′′ (Sup. Fig. 1b) was not associated with any natural partitioning in the newly reconstructed substitution matrix. It is important to note that splitting either serine, arginine or leucine into ii substitution groups, has a negligible effect on the commutation patterns of all other amino acids (Sup. Fig. S1).
In this mode we find that considering serine codon usage divides serine into two substitution groups. Based on the different properties of the serine substitutions groups, we postulated that the neutral amino acids that accept a propensity towards β-turns (GS′′North)7 would exist preferred at positions where the diversity of permissible amino acid usage is loftier, while the generally neutral amino acids (Due south′PA) would be prevalent in more conserved positions. The underlying rationale stems from the dominant role of β-turns in contact regions of receptors. Exchange among amino acids that share a propensity to form β-turn is most likely to change contact parameters while preserving the potential of the region to engage in binding and protein-protein interaction.
The distinct patterns of serine substitutions correlate with amino acid sequence variety
To test the second function of our hypothesis, that the two distinct serine commutation patterns volition be differentially expressed in conserved and diversifying amino acid positions, we assessed the correlation between the amino acid diversity and the fraction of amino acids occupied by the two substitution serine groups: GS′′N and South′PA. The analysis covered all positions in the human proteome where serine was ″meaningfully″ nowadays within multiple alignments of 99 vertebrate genomes (represented in UCSC Genome Browser Database9, see Methods). Using the measure of multifariousness10, we considered serine to be meaningful at a position if it was abundant enough to contribute to the diverseness of the sample, at that position10 (see Methods). Focusing at positions in which serine (Southward′ and/or S′′) was a meaningful amino acidx (see Methods) nosotros establish that the fraction of GS′′N usage at each sequence position showed a weak but significant positive correlation to variety of amino acids at that position while the fraction of S′PA usage showed a negative correlation (Spearman's rank correlation for GS″Due north rho = 0.146, p-value < 0.001, and for South′PA rho = −0.126, p value < 0.001). Inspecting more explicitly the summit and lesser quartiles of the diverseness distributions at different positions, nosotros plant the skew in the substitution patterns to be fifty-fifty more pronounced. We compared the levels of GS′′N and South′PA in the summit and lesser quartiles of positions ranked according to their degree of diverseness. We constitute that the fraction of GS′′North in near diverse positions was 6 folds more prevalent compared to the least diverse positions (median fraction GS′′N = 0.0588 vs. 0.0103). Based on the aforementioned assay, the fraction of S′PA shows the opposite trend, albeit to a lesser extent (median fraction S′PA = 0.131 vs. 0.141). In both cases these differences are pregnant (Mann-Whitney U examination: GS′′N p value < 0.001, S′PA p value < 0.001) (Fig. 2c).
GS′′N is over utilized in the fast diversifying regions of HIV envelope glycoprotein gp120
We next tested the possibility that the segmentation of S′ and Southward′′ was not only associated with conservation at the level of long-range evolution taxonomy but also associated with high diversity positions that govern poly peptide interaction. We tested the HIV envelope gp120, an exposed glycoprotein on the surface of the HIV envelope which is essential to virus entry into cells. The protein gp120 is a central determinant in the virus' ability to bind surface receptors, including CD4, that lead to viral fusion11,12. It is therefore a probable field of study for strong evolutionary constraints. At the same fourth dimension, gp120 is a prominent antigen that undergoes rapid adaptation to evade the allowed organisation13,fourteen. Information technology is thus an ideal showcase of a protein sequence comprised of amino acrid positions that are highly conserved and others that are hypervariable. Comparing the substitution patterns in these two types of positions, we considered only positions in which serine (S′ and/or South′′) was a meaningful amino acid (see Methods). The analysis was performed on a dataset of 4173 gp120 sequencesii (see Methods). Similarly, to the observations from the data set of vertebrate conservation (Fig. 2c), we found that GS′′N is over utilized in the fast diversifying regions of gp1202,3,4 (referred to as hotspot, median = 0.426), compared to the rest of the protein (median = 0.09) (Isle of mann-Whitney U test: p value = 6.11 × 10 −6 ). At the same time, S′PA shows a significant subtract in representation betwixt hotspots (median = 0.06) and the rest of the protein (median = 0.152) (p value = 0.04). (Fig. 3a and Sup. Fig. S2).
GS′′N and S′PA biases between regions of differing diversity. (a) Fraction of GS′′Northward (left) or South′PA (right) amino acids in HIV various hotspot regions (cherry-red) vs. other more conserved positions (blue). The distributions are represented by a box plot overlaying a violin plot. (b) Looking beyond clone populations in 40 humans we show (i) the distribution of the difference per private of the median fraction of clones that take more GS′′North (out full amino acids) in CDR compared to FWR germline positions and the median fraction of clones that have more GS′′N in FWR compared to CDR positions, where serine is meaningful (crimson); (two) the distribution of the difference per individual of the median fraction of clones that have more S′PA (out total amino acids) in CDR compared to FWR germline positions and the median fraction of clones that have more South′PA in FWR compared to CDR positions, where serine is meaningful (blue). (c) Distribution across individuals of the difference between median fraction of clones that have a higher fraction of GS′′N (out of GS′′Due north + S′PT) at a CDR somatic exchange position and the median fraction of clones that take a higher fraction of GS′′N (out of GS′′Due north + S′PT) at FWR somatic exchange positions.
A choice footprint in both germline and somatic B cell receptor populations
As our final example of the different commutation patterns of serine in diversifying and conserved positions, we considered both the somatic and germline substitution patterns in B jail cell receptors. Allow us first give a short introduction of this system to country the foundation for B cell receptors as a model of evolutionary selection process, emphasizing the special nature of somatic B cell receptor diversification. The somatic selection process is especially informative as we can elucidate its germline source and thus rails not just substitution patterns but too, and uniquely, their amino acrid source. B cells undergo two stages of differentiation and selection of their B prison cell receptors. Both stages are essential for the creation of a varied immune repertoire to recognize and fight diseases. In the first stage, B cells located in the bone marrow recombine germline Five(D) and J cistron segments3,five,six to create heavy and light chains which, when joined, class a unique B cell receptor3,6,7,15. B cells that grade a functional receptor in this fashion are selected and proliferate, producing B jail cell clones, each with a common progenitor and a common receptor type. During the immune response, the naive B jail cell population undergoes an additional process of affinity maturation. B cells proliferate, quickly mutate their B jail cell receptor genes and dice, such that the B cells with B cell receptors of higher binding to the antigens of the disease boss the B jail cell population. Since nearly of the human V, D, and J germline gene segments that recombine to encode the B cell receptor are knownvii,16, nosotros are able to assign every observed mutant B cell receptor sequence to its germline origin. In this way, we can identify clonotypes of sets of mutant B cells with common progenitors and characterize the precise types of surviving substitutions forth with their amino acid sources. Moreover, we tin can determine for every amino acrid in a sequence if it is germline encoded or the consequence of a somatic substitution during an allowed response. Finally, the B cell receptor, much like the HIV gp120, is too divided into highly variable complementarity determining regions (CDRs) and more than constrained framework regions (FWRs)9,17. The CDRs interact with antigens and therefore demand to rapidly arrange and diversify and are thus under diversifying selectionten,18,19,twenty. On the other hand, the FWRs serve as the backbones of the receptor. As such, these regions are constrained to maintain the rigidly of their construction, and thus are mostly under purifying selection10,xviii,21.
The ii serine exchange patterns differ in their prevalence in the variable and conserved positions of the B cell receptor repertoire
We analyzed the B cell repertoires of forty homo individuals from three different geographical location across the globe10,22,23,24 (run across Methods). For each private we characterized the full prepare of heavy chain sequences belonging to the B cell receptor populations. The repertoires of sequences were divided into clones, as described in a higher place (and encounter Methods). Focusing on the Five gene segment of the B cell receptor, whose germline is conspicuously defined, we annotated the unique positions in each clone which had either undergone an amino acid substitution or remained encoded for their germline amino acids. We only considered the positions in which serine (South′ or S′′) was a meaningful amino acid based on the diversity of the germline positions (come across Methods). Such positions were divided co-ordinate to their positions into those found in the hypervariable CDRs and the conserved FWRs8,20,25.
For each position we counted the number of clones in an private in which it was germline encoded and either GS′′Due north or S′PA. We so calculated the median level of GS′′Northward and S′PA usage in each region (CDR or FWR) across the serine positions. To assess whether the CDR utilized more than GS′′N amino acids with respect to FWR, we performed a Wilcoxon signed-rank tests beyond all xl individuals comparing individual median level of the departure of GS′′N in both B cell receptor regions (i.e. CDR minus FWR). We and then repeated the same test on Southward′PA levels across all 40 individuals. When considering germline positions, we constitute quite clearly and significantly that GS′′N was over abundant in the CDR of individuals while South′PA was over abundant in the FWR (Wilcoxon signed-rank test shows successes for GS′′N at 39 out of 40, p =7.91 × 10 −nine and S′PA in only 1 out of 40, p = 9.09 × x −12 ) (Fig. 3b and Sup. Fig. ii).
Somatic mutation and selection processes is skewed towards high affluence of GS″Northward in the highly variable CDR regions
Having established that germline positions of the B cell receptor genes showroom a preference for GS′′N in the hypervariable (CDRs) regions and Southward′PA in the more conserved ones (FWRs), we now asked whether such bias also revealed itself for the somatic mutations that signify substitutions with shorter timescale and loftier level of mutations. To this end, nosotros compared GS′′N somatic substitutions out of the total GS′′N + S′PA substitutions, for each individual with success existence when the fraction of GS′′Northward CDR to the fraction GS′′N FWR is positive (i.e.>0). This test is based on the notion that equally FWR regions more often than not do non show substitutions. We found that in the somatic exchange also, the aforementioned blueprint holds. Specifically, GS′′N is over abundant in the CDR compared to FWR (Wilcoxon signed-rank test reported a success for 27 out of 39, p =six.52 ×ten −5 ) (Fig. 3c and Sup. Fig. 4).
S″ phosphorylation sites are more prone to substitutions relative to those encoded by S′
To direct show a possible functional effect to the use of S′ or S″ and their permissible substitutions, we focused on the unique capacity of a subset of serine residues to be modified past serine/threonine kinases. We hypothesized that the tendency of phosphorserine exchange will be biased according to the type of serine encoding sets26. In all eukaryotes, serine (S), threonine (T) and tyrosine (Y) are the only amino acids that tin exist modified past kinases. To exam whether in that location is a deviation in the substitution patterns of S′ and Due south″ in the context of phosphorylation sites, we considered the coding variations from the human Exome Aggregation Consortium (ExAC) dataset. The ExAC dataset aggregates exome polymorphic sites of 60,706 unrelated healthy individuals. Altogether most eight.3 1000000 variations across the human population are reported, many occur at extremely low frequency (i.e. reported for a single individual). Nosotros mapped on the ExAC coding exomes 37,565 experimentally observed phosphorylated sites, among them 30,219 are phosphoserine (come across Methods). We reanalyzed the ExAC data and quantified the tendency of phosphoserine (p-S) to exhibit substitutions that could maintain its phosphorylation chapters when encoded past S′ or by S″ codon sets. To this end, we first congenital a substitution model that is based on the neutral model from all reported ExAC variations affecting the third position of the 4-fold degenerate amino acid codons (covers valine, proline, threonine, alanine and glycine, see Methods and Sup. Fig. v). Looking across all phosphorylation sites, nosotros compared the observed substitutions following single bespeak mutations to the expected pattern of the 4-fold degenerate model. Equally seen in Fig. 4, a single bespeak mutation from South′ and Southward″ would lead but to cysteine and threonine. Manifestly, a modify to cysteine would necessarily leads to a loss of a phosphosite, and this is every bit selected against from both S′ and S″ when compared to the l mutation model based on the 4-fold degenerate amino acids. Nonetheless, nosotros found a strong bias in the substitutions to threonine (T) from Due south′ or S″. Notably, substitution from serine to threonine is likely to maintain the phosphorylation potential due to the overlap in S/T kinases specificity27. While the S′ to T exchange is negatively selected against compared to the iv-fold degenerate mutation model, substitutions from Southward″ to T are not. The S′ codons are 32 folds less decumbent to alterations that retain phosphorylation capacity relative to Southward″ (Fig. 4). This result is in accord with our ascertainment for the not-randomized alternative positioning of S′ and Due south″ in the genome. The bias confronting South′ to T substitutions is not symmetrical28, we plant that the threonine phosphorylation (p-T) sites have apparently an equal frequency of substitutions to both S′ and Southward″ (Fig. 4, meridian). When we extended our analysis to all unmarried point mutations in any advent of Southward, T and Y across the ExAC, we found that in contrast to our ascertainment regarding the unique bias of functional p-S sites, the substitutions patterns of unmarried point mutations could be primarily explained by the 4-fold degenerate model of mutations (Fig. 4, bottom).
Serine phosphorylation sites evidence differences in conservation depending on codon usage. Substitution network based on single signal mutations based on the aggregation of human population polymorphism from the ExAC database. The tendency of substitution for South, T and Y for all phosphorylated positions (Superlative). All Due south, T and Y sites in the man proteome (Bottom). Arrows indicate the substitution directionality. The color of the pointer captures the relative abundance of the substitutions compared to the expected patterns of mutation equally calculated directly from the mutations in the third codon positions of 4-fold degenerate amino acids. Substitutions less arable than expected with a ratio < −1 (blue) ; substitutions more than abundant than expected with a ratio > ane (reddish) and substitutions that are at a similar degree as expected (i.east., −1 < ratio < 1) (gray). Values are rounded to testify the log ability of the substitution abundance. The verbal range of the relative abundance of all substitutions from S, Y and T to any other amino acid is shown next to the network view.
Our results regarding p-S substantiate the notion of a differential selection pressures which are associated with serine encoded past Due south′ or Southward″ codon sets. In the case of p-S, we observed that S′ sites are indicative by a reduced trend to be altered relative to Due south″, which is in accord with a S′ being engaged in a more conservative commutation patterns when compared to Due south″.
Give-and-take
In the results above nosotros have characterized patterns of substitution of serine both at the whole genome level under long-range evolution comparing the human genome to 99 other species, and at the human population level, analyzing protein substitutions from over 60,000 salubrious individuals. Nosotros further tested our hypothesis by focusing on two specific biological contexts, the somatic changes for B-cell receptors and gp120 an envelop gene of HIV. In both contexts, the genes include in single polypeptide chain distinguishable regions that are subjected to diversifying (positive) selection and other regions subjected to potent evolutionary constraints (purifying selection). Across all our examined datasets, we found a clear segregation of amino acid substitutions that are predicted past the division of serine encoding according to the genetic code. We show that the bias in serine codon usage previously found in B prison cell receptor repertoires29 has a role in maintaining variety beyond the immune B prison cell receptor repertoire. Indeed, it underlies a more than general segregation in amino acid substitution patterns that divides serine commutation into two groups linked to the diversity and functionality of cistron products. The commencement group (GS″N), mostly conserve for β-turns, are constitute in protein regions discipline to diversifying pick (east.k., protein contact regions). In contrast, the 2nd prepare (South′PA) comprise of more more often than not neutral amino acids and are institute in conserved protein regions, field of study to stronger evolutionary constraints. To show that South′ is under stricter purifying selection from a more functional perspective we looked also at the substitution patterns of p-S sites in the homo population (from ExAC dataset). We showed that while the majority of the phosphorylation sites in the human proteome are p-S (80.4%) of which ~sixty% are encoded by the S′ codon set. Still, across all p-S sites we found that substitutions from S′ showed a substantial negative selection to threonine, while no such pick is observed for serine that are encoded by the S″ codon set up (Fig. 4).
Determination
Nosotros have thus shown that in biological pick processes the codons of serine indicate different types of choice for the amino acid and its permissible substitutions. We have shown the importance of this special characteristic of serine, in general and for phosphorylation sites, beyond multiple scales of evolutionary selection: beyond species, within human population and for the somatic B prison cell selection and viral quasi species. At all these scales of selection the S′ codon set up is under a stronger purifying selection while S″ codon set tends to undergo diversifying selection, every bit is reflected from protein sequence, structure and function.
Based on the cumulative observations from vertebrates and human-centric evolution, allowed and viral option nosotros notice that in highly diversified positions of amino acids, when serine is present, it is more often encoded by AGY and will substitute in addition to whatsoever synonymous changes to glycine and asparagine (GS″North). In contrast, in highly conserved positions serine is more than frequently encoded past TCN and will tend to substitute in a non-synonymous form to proline and alanine (Southward′PA). In this way nosotros prove that codon usage and not but amino acid type serves as an indicator of selection. We provide a farther back up for the view that the genetic code has evolved to permit maintenance of several types of substitution patterns.
Materials and Methods
Assay of whole genome exon sequences
Sequence collection
All 162,633 known canonical exon gene sequences of all isoforms from the multiple alignments of 99 vertebrate genomes with human dataset were obtained from the UCSC Genome Browser database9,xiii,14. A list of each vertebrate used by UCSC, along with their methodology and parameters, can be found at http://hgdownload.soe.ucsc.edu/goldenPath/hg38/multiz100way/.
BLOSUM analysis
BLOSUMs were created using our ain implementation of the BLOSUM algorithm2,viii in guild to account for treating S′/Southward′′, Fifty′/50′′ and R′/R″ differently. The code is open source and available at https://github.com/GregorySchwartz/blosum
Calculation of by position multifariousness of amino acids
For each gene isoform in the data set, the diversity of amino acids was calculated at each position. In order to avoid bias of highly arable or rare amino acids, the measurements were taken with an gild of onethirty. The calculated per position diversity was used to estimate the meaningful types of amino acids found at each position. At each position, only the n most abundant types of amino acids were included, where northward is the amino acid diversity at that position. Nosotros defined meaningfully nowadays serine positions as those in which serine was one of its n most abundant amino acids10,nineteen. Positions were gathered into iv groups based on the quartiles of multifariousness, with the high diversity positions having diversities greater than the upper quartile and low diversity positions having diversities less than the lower quartile, excluding outliers.
GS′′N and South′PA comparisons
Within the amino acrid positions where serine was a meaningful amino acid, the abundance of GS′′N and S′PA, normalized by the number of sequences at each position across all sequences (the ″fraction″ of each category at a position), were compared in the high diversity and depression variety positions. Spearman'southward rank correlation was used to summate the correlation of GS′′Due north or Due south′PT fractions with diverseness. All comparisons between the upper and lower multifariousness positions and between GS′′N and S′PA were washed using the Mann-Whitney U 2-tailed test.
Analysis of gp120
We collected 4173 HIV-1 env gene sequences filtered by alignment to HXB2 HIV reference genome from the Los Alamos National Laboratory HIV Database (http://www.hiv.lanl.gov/)two and analyzed their gp120 region. We considered all 145 positions in which S (S′ and/or S′′) was a meaningful amino acrid, defined every bit in the known exon analysis above. Within these amino acrid positions, the abundance of GS′′N and S′PA was normalized by the number of sequences at each position beyond all 4173 sequences. We then compared the abundance of the two substitution groups in the 47 hypervariable positions2 and in the 98 remaining positions (Sup. Fig. S3). Following the above results significance of all comparisons was calculated using the Isle of man-Whitney U ane-tailed test.
Analysis of the heavy chain B cell receptor
Sequence collection
Heavy chain B cell receptor sequences were gathered from iv different groups of individuals in dissimilar populations: (1) Peripheral claret samples from three immature healthy individuals (D1)22; (2) 6 young (D2 young) and vi old (D2 old) patients sampled at immunization to influenza and seven or 28 days after23; (3) 14 Papua New Republic of guinea private repertoires (D3)24, and (4) 12 Australian individual repertoires (D4)24. Samples D1 and D2 were sequenced using the Roche 454 sequencing technology (FLX Titanium)22,23, while D3 and D4 where Sanger sequenced on an Applied Biosystems (ABI 3730) auto24. Since individuals and samples were compared, individuals that had < iii sequences were removed. Therefore, D3 01 in the D3 data set24 was removed.
Clonal definition and mutation counts
In all cases, all sequences were locally aligned using Smith-Waterman with a gap penalty of xvi to conform to the ImMunoGeneTics database (IMGT) definitions and position numbering25. To these we added aboriginal germlines to account for alleles evolved in New Guinea31. But the Five gene part of the gene was analysed (to amino acid position 106). The CDR and FWR were modified IMGT definitions that were used in previous analysis: FWR1 = 1–24; CDR1 = 25–40; FWR2 = 41–53; CDR2 = 56–65; FWR3 = 66–104; CDR3 = 105–106320,25. To avoid possible cryptic germline V gene assignments, sequences were removed if they were with over 30% of their nucleotides being mutated. This filtering resulted in 1336 sequences from D1, twenty,102 sequences from D2, 1098 sequences D3, and 636 sequences from D4, resulting in data from xl individuals.
The sequences were separated into clones identified past having the aforementioned V gene, J gene, and CDR3 length. This resulted in ane,290 clones from D1, nine,391 clones from D2, 699 clones from D3, and 231 clones D4. The exact number of positions sequences in the unlike data sets varies. Therefore, in order to have each amino acrid sequence position represented in most if non all clones, positions represented in less than 30 sequences in whatever of the data sets were excluded. Thus all V genes were analyzed merely in the post-obit IMGT positions: 25–xxx, 35–59, 63–72, 74–10625 Inside each clone, each mutational result was counted only once. A mutational upshot in the recombined information sets was defined equally ane nucleotide change in a codon.
Calculation of by position variety of amino acids
For each private in each data fix, nosotros separately calculated the diversity of germline amino acids and the diversity of amino acids substituted from their respective germline at each position across clones.
GS′′N and Due south′PA comparisons
GS′′Northward and/or Due south′PA levels amongst somatically substituted and germline maintained amino acids in each position in each private, were compared only in the 27 positions in which S (Due south′ and/or South′′) was a meaningful amino acid in the germline-maintained positions. Meaningful amino acids were again defined as in the known exon analysis described in a higher place. In these positions (marked in cherry-red in Sup. Fig. 4), the abundance of GS′′N and Due south′PA were compared, normalized by the number of ″unique″ instances of amino acids at each position inside the CDRs and the FWRs. Only 26 of the 27 positions had substitutions and therefore the comparing of substituted positions was only done on these 26 positions. The comparisons were done by comparing the medians of GS′′N and Southward′PA distributions beyond positions in the CDRs and FWRs using the Wilcoxon Signed Rank test. Individual A12 was not nowadays in this analysis of the somatically substituted BCR sequence positions equally we did not observe any substitutions in our 27 positions of interest in the BCR sequences of that private.
Analysis of phosphorylation sites from ExAC database
The Exome Aggregation Consortium (http://ftp.broadinstitute.org/pub/ExAC_release/release0.3.1/) (ExAC) combines the genetic variation observed in 60,706 unrelated healthy individuals. All observed single nucleotide variations (SNVs) are recorded for each position in the genome, along with the frequency of chromosomes that display this SNVs, and its quality score. The dataset contains an average of 12.5 SNVs per 100 nt in the exome. The Genome Reference Consortium Human Build 37 (GRCh37, hg19) was used for edifice this VCF version. Nosotros only considered high quality SNVs that are within a canonical CDS. Stop codons were not considered inside the CDS. These filtrations left u.s. with 89.7% of the original data. SNVs were translated to single codon variations with respect to their position in the genome, using the our Geneffect Variant Consequence Predictor (https://github.com/nadavbra/geneffect). Geneffect as well provided a user-friendly interface for extracting sequence characteristic annotations from UniProtKB32. Specifically, out of 1,386,569 serine residues in hg19, a total of 30,219 sites were reported as phosphoserines according to Sequence features from UniProtKB. Based on these information, a human specific codon substitution matrix, was synthetic. Each matrix jail cell represents the observed frequency of the substitution of a row codon past its column codon. The substitution of the major to the minor alleles is considered. A normalization was performed by the number of appearances of each codon in the canonical CDSs. This produced a 61 × 61 substitution matrix, several of the matrix cells are unoccupied due to the request for multiple steps for amino acid exchange. The diagonal entries, which represent the probability for a not-exchange, completed each row sum to 1. As a reference for neutral mutations, we created a 4-degenerate matrix, which considers merely mutations that appear in the third base of 4 degenerate codons (it comprises ~86.5% of all mutations). The degenerate matrix was then converted to a nucleotide resolution, translating into a 4 × iv mutation matrix. This four-degenerate nucleotide matrix was translated dorsum to a codon substitution 4-degenerat matrix. This is the joint probability of the single nucleotide mutations assuming independence of all three positions inside a codon. For phosphorylation exam we included experimentally validated phosphoserine (xiii,092 sites), phosphothreonine (3,326) and phosphotyrosine (873 sites) based on the Sequence features from UniProtKB.
References
-
Rogozin, I. B. et al. Evolutionary switches between 2 serine codon sets are driven past selection. Proceedings of the National Academy of Sciences 113, 13109–13113 (2016).
-
Foley, B. et al. HIV Sequence Compedium 2015. hiv.lanl.gov (2015). Available at: http://world wide web.hiv.lanl.gov/content/sequence/HIV/COMPENDIUM/2015/sequence2015.pdf. (Accessed: xvi September 2016)
-
Hershberg, U. & Shlomchik, G. J. Differences in potential for amino acrid modify following mutation reveals distinct strategies for κ and λ calorie-free concatenation variation. Proc. Natl. Acad. Sci. USA 103, 15963–15968 (2006).
-
Fischer, W. et al. Singled-out Evolutionary Pressures Underlie Diversity in Simian Immunodeficiency Virus and Human Immunodeficiency Virus Lineages. Periodical of Virology 86, 13217–13231 (2012).
-
Schatz, D. One thousand. & Ji, Y. Recombination centres and the orchestration of 5(D)J recombination. Nat Rev Immunol 11, 251–263 (2011).
-
Chothia, C., Gelfand, I. & Kister, A. Structural determinants in the sequences of immunoglobulin variable domain. J Mol Biol 278, 457–479 (1998).
-
Chou, P. Y. & Fasman, Yard. D. Prediction of beta-turns. Biophysical Journal 26, 367–383 (1979).
-
Henikoff, S. & Henikoff, J. M. Amino acid substitution matrices from poly peptide blocks. Proceedings of the National University of Sciences 89, 10915–10919 (1992).
-
Rosenbloom, One thousand. R. et al. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 43, D670–D681 (2015).
-
Schwartz, G. Westward. & Hershberg, U. Conserved variation: identifying patterns of stability and variability in BCR and TCR V genes with different diversity and richness metrics. Phys Biol 10, 035005 (2013).
-
Lasky, L. A. et al. Delineation of a region of the human immunodeficiency virus type ane gp120 glycoprotein critical for interaction with the CD4 receptor. Cell l, 975–985 (1987).
-
Smith, D. et al. Blocking of HIV-1 infectivity by a soluble, secreted course of the CD4 antigen. Science 238, 1704–1707 (1987).
-
Wyatt, R. et al. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393, 705–711 (1998).
-
Novitsky, V., Lagakos, S., Herzig, G. & Bonney, C. Evolution of proviral gp120 over the first year of HIV-ane subtype C infection. Virology (2009).
-
Tonegawa, S. Somatic generation of antibody multifariousness. Nature 302, 575–581 (1983).
-
Lefranc, M. P. et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 593–597 (2005).
-
Wu, T. T. & Kabat, E. A. An assay of the sequences of the variable regions of Bence Jones proteins and Myeloma light chains and their implications for antibiotic coplementarity. The Journal of Experimental Medicine 132, 211–250 (1970).
-
Anderson, S. Thousand. et al. Taking advantage: loftier-affinity B cells in the germinal heart have lower death rates, but similar rates of division, compared to low-affinity cells. J. Immunol. 183, 7314–7325 (2009).
-
Schwartz, M. W. & Hershberg, U. Germline amino acid variety in B cell receptors is a good predictor of somatic choice pressures. Frontiers in Immunology 4 (2013).
-
Kabat, E. A., Wu, T. T., Reid-Miller, Thou., Perry, H. & Gottesman, Chiliad. Sequences of proteins of immunological involvement. (US Govt. Printing Off. No. 165–492., 1987).
-
Shlomchik, M. J., Aucoin, A. H., Pietsky, D. S. & Weigert, G. G. Construction and function of anti-Dna autoantibodies derived from a single autoimmune mouse. Proc. Natl. Acad. Sci. 84, 9150–9154 (1987).
-
Wu, Y.-C. et al. High-throughput immunoglobulin repertoire analysis distinguishes between human IgM retentivity and switched retentiveness B-prison cell populations. Blood 116, 1070–1078 (2010).
-
Wu, Y.-C., Kipling, D. & Dunn-Walters, D. M. Age-related changes in human being peripheral blood IGH repertoire following vaccination. Frontiers in Immunology 3, 1–12 (2012).
-
Wang, Y. et al. IgE Sequences in Individuals Living in an Area of Endemic Parasitism Show Little Mutational Evidence of Antigen Selection. Scandinavian Journal of Immunology 73, 496–504 (2011).
-
Alamyar, Due east., Giudicelli, V., Li, Southward., Duroux, P. & Lefranc, M.-P. IMGT/HighV-QUEST: the IMGT® web portal for immunoglobulin (IG) or antibody and T prison cell receptor (TR) analysis from NGS high throughput and deep sequencing. Immunome Enquiry 8, 26–26 (2012).
-
Creixell, P., Schoof, Due east. G., Heng Tan, C. Due south. & Linding, R. Mutational properties of amino acid residues: implications for evolvability of phosphorylatable residues. Philosophical Transactions of the Regal Society B: Biological Sciences 367, 2584 (2012).
-
Chen, C. et al. Identification of a Major Determinant for Serine-Threonine Kinase Phosphoacceptor Specificity. Molecular Cell 53, 140–147 (2014).
-
Chen, South. C. C., Chen, F. C. & Li, West. H. Phosphorylated and Nonphosphorylated Serine and Threonine Residues Evolve at Unlike Rates in Mammals. Mol Biol Evol 27, 2548–2554 (2010).
-
Wagner, S. D., Milstein, C. & Neuberger, 1000. Due south. Codon bias targets mutation. Nature 376, 732 (1995).
-
Jost, Fifty. Entropy and diverseness. Oikos 113, 363–375 (2006).
-
Wang, Y. et al. Genomic screening past 454 pyrosequencing identifies a new human IGHV cistron and xvi other new IGHV allelic variants. Immunogenetics 63, 259–265
-
Brandes, N., Linial, N. & Linial, Thousand. Quantifying gene pick in cancer through poly peptide functional alteration bias. Nucleic Acids Res. 458, 719 (2019).
Acknowledgements
Gregory Due west. Schwartz was funded past the U.S. Department of Didactics Graduate Assistance in Areas of National Demand (GAANN) program, CFDA Number: 84.200. The authors would like to give thanks Nadav Brandes for his support in the ExAC analysis and providing the Geneffect platform. The authors would like to thank Ruth Hershberg for fruitful discussions and turns of phrase and Edward Trifonov for the conversation that started this report.
Author information
Affiliations
Contributions
T.S. and One thousand.L. designed the analysis of ExAC data. M.Fifty. and U.H. interpreted the results. G.W.S. and U.H. designed the rest of the analysis and interpreted their results. One thousand.W.Due south., Grand.Fifty. and U.H. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional data
Publisher'southward note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Admission This article is licensed under a Creative Commons Attribution iv.0 International License, which permits use, sharing, adaptation, distribution and reproduction in whatever medium or format, as long as yous give advisable credit to the original author(south) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other tertiary party material in this commodity are included in the article's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is non included in the article's Artistic Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a re-create of this license, visit http://creativecommons.org/licenses/by/4.0/.
Reprints and Permissions
Virtually this commodity
Cite this article
Schwartz, G.W., Shauli, T., Linial, Yard. et al. Serine substitutions are linked to codon usage and differ for variable and conserved protein regions. Sci Rep 9, 17238 (2019). https://doi.org/10.1038/s41598-019-53452-three
-
Received:
-
Accustomed:
-
Published:
-
DOI : https://doi.org/10.1038/s41598-019-53452-3
Comments
Past submitting a comment you concur to abide by our Terms and Customs Guidelines. If you discover something abusive or that does non comply with our terms or guidelines please flag it every bit inappropriate.
Source: https://www.nature.com/articles/s41598-019-53452-3
0 Response to "mutation from threonine to what amino acid make phosphorylation"
Post a Comment